


Next: Using Algorithm5 for classification
Up: Structural Alignment algorithm based
Previous: Extending Algorithm1 for atomic
Computing the actual Local structural alignment
With all the relevant background at this stage we now describe our
algorithm which finds highly similar sub-structures between two proteins
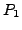 and
and 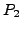 for details on how the atomic coordinates in
for details on how the atomic coordinates in 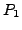 and
and 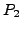 are described please refer to section3.1. And let
are described please refer to section3.1. And let
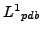 and
and
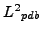 be the linear list of atoms as described in
section3.1,the details of our algorithm can be summarized
as follows.
be the linear list of atoms as described in
section3.1,the details of our algorithm can be summarized
as follows.
- Select a window of size
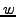 , slide it across the list's
, slide it across the list's
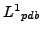 and
and
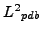 . Let
. Let 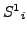 be the atoms from
be the atoms from
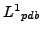 at a position
at a position 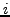 in list
in list
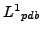 , similarly define
, similarly define 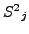 for atoms within the window at position
for atoms within the window at position 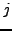 in
in
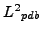 .
.
- For every such
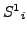 ,
, 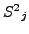
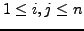 , compute
, compute
![$ {V^1}[i]$](img67.png) ,
, ![$ {V^2}[j]$](img68.png) , which are the same vectors in Algorithm1
, which are the same vectors in Algorithm1
- Define a cost matrix
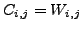 , where
, where 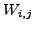 is
the weighted distance (defined in section3.4) between
vectors
is
the weighted distance (defined in section3.4) between
vectors ![$ {V^1}[i]$](img67.png) and
and ![$ {V^2}[j]$](img68.png) (defined in above step each of size
(defined in above step each of size 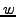 ).
).
- Use a variation of our dynamic programming(see Algorithm4 we will explain this in the next section)algorithm, the algorithm is similar to Longest Common Substring (Please note the difference between Substring and Subsequence both are different problems).
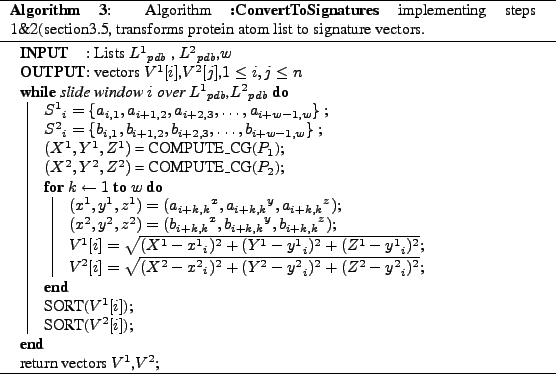
Now we give algorithms corresponding each of the steps described steps 1&2 are done by Algorithm3, steps 3&4 are performed by Algorithm4. Finally we put all these into a
high-level Algorithm5 for better understanding.
boxed
boxed

boxed
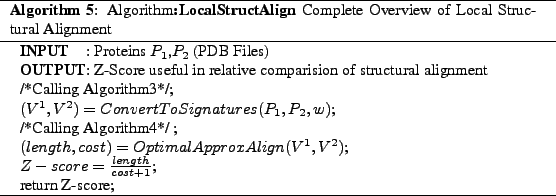
We have given the details of our Local Structural Alignment algorithm
which starts with LocalStructAlign (Algorithm5)
and produces a metric called the Z-Score we use this metric in
finding the closest structurally similar proteins. In the next section
we describe how we use the Z-Score computed by Algorithm5 in classification of the protein into classes and super families.We also describe on how we used Algorithm5 in finding
structural motifs in section5.



Next: Using Algorithm5 for classification
Up: Structural Alignment algorithm based
Previous: Extending Algorithm1 for atomic
Vamsi Kundeti
2007-10-10
